MAKER Tutorial
Contents
- 1 About MAKER
- 2 Introduction to Genome Annotation
- 3 MAKER Overview
- 4 Installation
- 5 Getting Started with MAKER
- 6 Details of What is Going on Inside of MAKER
- 7 MAKER's Output
- 8 Viewing MAKER Annotations
- 9 Advanced MAKER Configuration, Re-annotation Options, and Improving Annotation Quality
- 9.1 Configuration Files in Detail
- 9.2 Training ab initio Gene Predictors
- 9.3 MAKER Web Annotation Service
- 9.4 mRNAseq
- 9.5 Improving Annotation Quality with MAKER's AED score
- 9.6 Merge/Resolve Legacy Annotations
- 9.7 MPI Support
- 9.8 User Interface for Local MAKER Instalation
- 9.9 MAKER Accessory Scripts
About MAKER
MAKER is an easy-to-use genome annotation pipeline designed for small research groups with little bioinformatics experience. However, MAKER is also designed to be scalable and is thus appropriate for projects of any size including use by large sequence centers. MAKER can be used for de novo annotation of newly sequenced genomes, for updating existing annotations to reflect new evidence, or just to combine annotations, evidence, and quality control statistics for use with other GMOD programs like GBrowse, JBrowse, Chado, and Apollo.
MAKER has been used in many genome annotation projects:
- Schmidtea mediterranea - planaria (A Alvarado, Stowers Institute) PubMed
- Pythium ultimum oomycete (R Buell, Michigan State Univ.) PubMed
- Pinus taeda - Loblolly pine (A Stambolia-Kovach, Univ. California Davis) PubMed
- Atta cephalotes - leaf-cutter ant (C Currie, Univ. Wisconsin, Madison) PubMed
- Linepithema humile - Argentine ant (CD Smith, San Francisco State Univ.) PubMed
- Pogonomyrmex barbatus - red harvester Ant (J Gadau, Arizona State Univ.) PubMed
- Conus bullatus - cone snail (B Olivera Univ. Utah) PubMed
- Petromyzon marinus - Sea lamprey (W Li, Michigan State) PubMed
- Fusarium circinatum - pine pitch canker (B Wingfield, Univ. Pretoria) - Manuscript in preparation
- Cardiocondyla obscurior - tramp ant (J Gadau, Arizona State Univ.) - Manuscript in preparation
- Columba livia - pigeon (M Shapiro, Univ. Utah) - Manuscript in preparation
- Megachile routundata alfalfa leafcutter bee () - Manuscript in preparation
- Latimeria menadoensis - african coelacanth () - PubMed
- Nannochloropsis - micro algae (SH Shiu, Michigan State Univ.) PubMed
- Arabidopsis thale cress re-annotation (E Huala, TAIR) - Manuscript in preparation
- Cronartium quercuum - rust fungus (JM Davis, Univ. Florida) - Annotation in progress
- Ophiophagus hannah - King cobra (T. Castoe, Univ. Colorado) - Annotation in progress
- Python molurus - Burmese python (T. Castoe, Univ. Colorado) - Annotation in progress
- Lactuca sativa - Lettuce (RW Michelmore) - Annotation in progress
- parasitic nematode genomes (M Mitreva, Washington Univ)
- Diabrotica virgifera - corn rootworm beetle (H Robertson, Univ. Illinois)
- Oryza sativa - rice re-annotation (R Buell, MSU)
- Zea mays - maize re-annotation (C Lawrence, MaizeGDP)
- Cephus cinctus - wheat stem sawfly (H Robertson, Univ. Illinois)
- Rhagoletis pomonella - apple maggot fly (H Robertson, Univ. Illinois)
Introduction to Genome Annotation
What Are Annotations?
Annotations are descriptions of different features of the genome, and they can be structural or functional in nature.
Examples:
- Structural Annotations: exons, introns, UTRs, splice forms (Sequence Onotology)
- Functional Annotations: process a gene is involved in (metabolism), molecular function (hydrolase), location of expression (expressed in the mitochondria), etc. (Gene Ontology)
It is especially important that all genome annotations include an evidence trail that describes in detail the evidence that was used to both suggest and support each annotation. This assists in curation, quality control and management of genome annotations.
Examples of evidence supporting a structural annotation:
- Ab initio gene predictions
- Transcribed RNA (mRNA-Seq/ESTs/cDNA/transcript)
- Proteins
Importance of Genome Annotations
Why should the average biologist care about genome annotations?
Genome sequence itself is not very useful. The first question that occurs to most of us when a genome is sequenced is, "where are the genes?" To identify the genes we need to annotate the genome. And while most researchers probably don't give annotations a lot of thought, they use them everyday.
Examples of Annotation Databases:
Every time we use techniques such as RNAi, PCR, gene expression arrays, targeted gene knockout, or ChIP we are basing our experiments on the information derived from a digitally stored genome annotation. If an annotation is correct, then these experiments should succeed; however, if an annotation is incorrect then the experiments that are based on that annotation are bound to fail. Which brings up a major point:
- Incorrect and incomplete genome annotations poison every experiment that uses them.
Quality control and evidence management are therefore essential components to the annotation process.
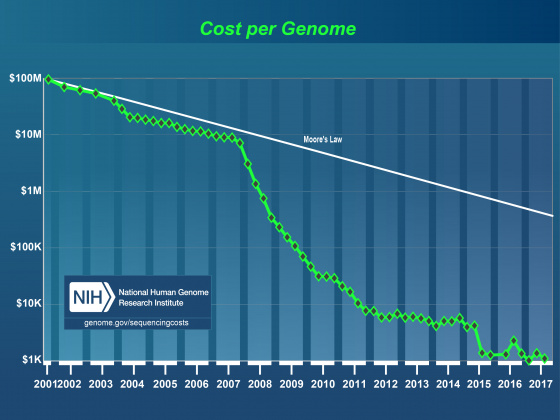
Effect of NextGen Sequencing on the Annotation Process
It’s generally accepted that within the next few years it will be possible to sequence even human sized genomes for as little as $1,000. As costs have drop, read lengths have increased, and assembly and alignment algorithms have matured the genome project paradigm is shifting. Even small research groups are turning their focus from the individual reference genome to the population. This shift in focus has already lead to great insights into the genomic effects of domestication and is very promising in helping us understand multiple host-pathogen relationships. Importantly these population-based studies still require a well-annotated reference genome. Unfortunately, advances in annotation technology have not kept pace with genome sequencing, and annotation is rapidly becoming a major bottleneck affecting modern genomics research.
For example:
- As of August 2012, 2,555 eukaryote and 11,099 prokaryote genome projects were underway.
- If we assume 10,000 genes per genome, that’s over 255,000,000 new annotations (with this many new annotations, quality control and maintenance is a major issue).
- While there are organizations dedicated to producing and distributing genome annotations (i.e ENSEMBL, JGI, Broad), the shear volume of newly sequenced genomes exceeds both their capacity and stated purview.
- Small research groups are affected disproportionately by the difficulties related to genome annotation, primarily because they often lack bioinformatics resources and must confront the difficulties associated with genome annotation on their own.
MAKER is an easy-to-use annotation pipeline designed to help smaller research groups convert the coming tsunami of genomic data provided by next generation sequencing technologies into a usable resource.
MAKER Overview
 The easy-to-use annotation pipeline.
The easy-to-use annotation pipeline.
| User Requirements: | Can be run by small groups (single individual) with a little linux experience |
|---|---|
| System Requirements: | Can run on desktop computers running Linux or Mac OS X (but also scales to large clusters and we're working on the iPhone App) |
| Program Output: | Output is compatible with popular GMOD annotation tools like Apollo, GBrowse JBrowse |
| Availability: | Free, open-source application (academic use) |
What does MAKER do?
- Identifies and masks out repeat elements
- Aligns ESTs to the genome
- Aligns proteins to the genome
- Produces ab initio gene predictions
- Synthesizes these data into final annotations
- Produces evidence-based quality values for downstream annotation management

What sets MAKER apart from other tools (ab initio gene predictors etc.)?
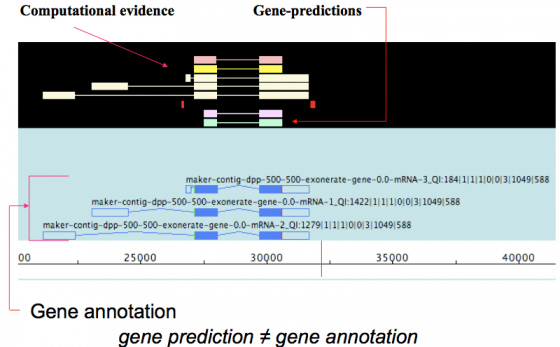
MAKER is an annotation pipeline, not a gene predictor. MAKER does not predict genes, rather MAKER leverages existing software tools (some of which are gene predictors) and integrates their output to produce what MAKER finds to be the best possible gene model for a given location based on evidence alignments.
gene prediction ≠ gene annotation
- gene predictions are partial gene models.
- gene annotations are gene models but should include a documented evidence trail supporting the model in addition to quality control metrics.
This may seem like a matter of semantics since the output for both ab initio gene predictors and the MAKER pipeline are conceptually the same - a collection of gene models. However there are significant differences that are discussed below.
Emerging vs. Classic Model Genomes
Not all genomes are created equal - each comes with its own set of issues that are not necessarily found in classic model organism genomes. These include difficulties associated with repeat identification, gene finder training, and other complex analyses. Emerging model organisms are often studied by small research communities which may lack the infrastructure and bioinformatics expertise necessary to 'roll-ther-own' annotation solution.
| 'Old School' Model Organism Annotation | 'New Cool' Emerging Organism Annotation |
|---|---|
|
Well developed experimental systems |
The genome will be a central resource for experimental design |
|
Much prior knowledge about genome/transcriptome/proteome |
Limited prior knowledge about genome |
| Large community | Small communities |
| $$$ | $ |
| Examples: D. melanogaster, C. elegans, human | Examples: oomycetes, flat worms, cone snail |
Comparison of Algorithm Performance on Model vs. Emerging Genomes
If you have looked at a comparison of gene predictor performance on classic model organisms such as C. elegans you might conclude that ab initio gene predictors match or even outperform state of the art annotation pipelines, and the truth is that, with enough training data, they do very well. It is important to keep in mind, however, that ab initio gene predictors have been specifically optimized to perform well on model organisms such as Drosophila and C. elegans, organisms for which we have large amount of pre-existing data to both train and tweak the prediction parameters.
What about emerging model organisms for which little data is available? Gene prediction in classic model organisms is relatively simple because there are already a large number of experimentally determined and verified gene models, but with emerging model organisms, we are lucky to have a handful of gene models to train with. As a result ab initio gene predictors generally perform very poorly on emerging genomes.
By using ab inito gene predictors within the MAKER pipeline you get several key benefits:
- MAKER provides gene models together with an evidence trail - useful for manual curation and quality control.
- MAKER provides a none|thumbwork within which you can train and retrain gene predictors for improved performance.
- MAKER's output (including supporting evidence) can easily be loaded into a GMOD compatible database for annotation distribution.
- MAKER's annotations can be easily updated with new evidence by passing existing annotation sets back though MAKER.
Installation
Prerequisites
Perl Modules
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN
- Template:CPAN(Optional, for accessory scripts)
- Template:CPAN(Optional, for accessory scripts)
- Template:CPAN(Optional, for MPI scripts)
- Template:CPAN(Optional, for MPI scripts)
External Programs
- Perl 5.8.0 or Higher
- SNAP version 2009-02-03 or higher
- RepeatMasker 3.1.6 or higher
- Exonerate 1.4 or higher
- NCBI BLAST 2.2.X or higher
Optional Components:
- Augustus 2.0 or higher
- GeneMark-ES 2.3a or higher
- FGENESH 2.6 or higher
Required for optional MPI support:
The MAKER Package
MAKER can be downloaded from:
- http://www.yandell-lab.org/ - but it should already be on the image
MAKER is already installed on the Amazon Machine Image that we will be using today, so let's start an instance of that AMI. Navigate your browser to:
The AWS EC2 Management Console
Search for the AMI named MAKER_2012-08-25_GMOD_03
Launch and instance of that AMI with the following parameters:
- Instance Type: Small (m1.small)
- Use a security group with:
- Allow SSH (port 22)
- Allow HTTP (port 80)
When the machine is running connect to it with SSH (puTTY):
Ensure you change the user name from root to ubuntu
Now that we're on our MAKER annotation server let's look at our MAKER installation:
cd /usr/local/maker ls -1
Note: That is a dash one, not a dash L, on the ls command.
You should now see the following:
bin data exe GMOD INSTALL lib LICENSE MWAS perl README src
There are two files in particular that you would want to look at when installing MAKER - INSTALL and README. INSTALL gives a brief overview of MAKER and prerequisite installation. Let's take a look at this.
less INSTALL
You shouldn't need to do this if MAKER is pre-installed or if MAKER installed all the prerequisites. But for the sake of documentation...
***Installation Documentation***
How to Install Standard MAKER
**IMPORTANT FOR MAC OS X USERS**
You will need to install developer tools (i.e. Xcode) from your installation
disk. If you are on a 32-bit system also install Rosetta from your instalation
disk. If you are using a 64-bit system, install fink (http://www.finkproject.org/)
and then install glib2-dev via fink (i.e. fink install glib2-dev). Make sure to
install fink as 64-bit when asked (32-bit is the default).
EASY INSTALL
1. Go to the .../maker/src/ directory and run 'perl Build.PL' to configure.
2. Accept default configuration options by just pressing enter.
3. type './Build install' to complete the installation.
4. If anything fails, either use the ./Build file commands to retry the
failed section or follow the detailed install instructions below to
manually install missing modules or programs. Use ./Build status to see
available commands.
./Build status #Shows a status menu
./Build installdeps #installs missing PERL dependencies
./Build installexes #installs missing external programs
./Build install #installs MAKER
Note: When using ./Build to install missig external programs, MAKER will use
the maker/src/locations file to identify download URLs. You can edit this file
to point at alternate locations.
DETAILED INSTALL
1. You need to have perl 5.8.0 or higher installed. You can verify this by
typing perl -v on the command line in a terminal.
You will also need to install the following perl modules from CPAN.
*DBI
*DBD::SQLite
*Proc::ProcessTable
*threads (Required by MPI and accessory scripts)
*IO::All (Required by accessory scripts)
*IO::Prompt (Required by accessory scripts)
*File::Which
*Perl::Unsafe::Signals
*Bit::Vector
*Inline::C
*PerlIO::gzip
a. Type 'perl -MCPAN -e shell' to access the CPAN shell. You may
have to answer some configuration questions if this is your first time
starting CPAN. You can normally just hit enter to accept defaults.
Also you may have to be logged in as 'root' or use sudo to install
modules via CPAN.
b. Type 'install DBI' to install the first module, then type
'install DBD::SQLite' to install the next, and so on.
c. Alternatively you can download moadules from http://www.cpan.org/.
Just follow the instructions that come with each module to install.
2. Install BioPerl 1.6 or higher. Download the Core Package from
http://www.bioperl.org
-quick and dirty installation-
(with this option, not all of bioperl will work, but what MAKER uses will)
a. Download and unpack the most recent BioPerl package to a directory of your
choice, or use Git to access the most current version of BioPerl. See
http://www.bioperl.org for details on how to download using Git.
You will then need to set PERL5LIB in your .bash_profile to the location
of bioperl (i.e. export PERL5LIB="/usr/local/bioperl-live:$PERL5LIB").
-full BioPerl instalation via CPAN-
(you will need sudo privileges, root access, or CPAN configured for local
installation to continue with this option)
a. Type perl -MCPAN -e shell into the command line to set up CPAN on your
computer before installing bioperl (CPAN helps install perl dependencies
needed to run bioperl). For the most part just accept any default options
by hitting enter during setup.
b. Type install Bundle::CPAN on the cpan command line. Once again just press
enter to accept default installation options.
c. Type install Module::Build on the cpan command line. Once again just
press enter to accept default installation options.
d. Type install Bundle::BioPerl on the cpan command line. Once again press
enter to accept default installation options.
-full BioPerl instalation from download-
a. Unpack the downloaded bioperl tar file to the directory of your choice or
use Git to get the most up to date version. Then use the terminal
to find the directory and type perl Build.PL in the command line, then
type ./Build test, then if all tests pass, type ./Build install. This
will install BioPerl where perl is installed. Press enter to accept all
defaults.
3. Install either WuBlast or NCBI-BLAST using instruction in 3a and 3b
3a. Install WuBlast 2.0 or higher (Alternatively install NCBI-BLAST in 3b)
(WuBlast has become AB-Blast and is no longer freely available, so if you
are not lucky enough to have an existing copy of WuBlast, you can use NCBI
BLAST or BLAST+ instead)
a. Unpack the tar file into the directory of your choice (i.e. /usr/local).
b. Add the following in your .bash_profile (value depend on where you chose
to install wublast):
export WUBLASTFILTER="/usr/local/wublast/filter"
export WUBLASTMAT="/usr/local/wublast/matrix"
c. Add the location where you installed WuBlast to your PATH variable in
.bash_profile (i.e. PATH="/usr/local/wublast:$PATH").
3b. Install NCBI-BLAST 2.2.X or higher (Alternatively install WuBlast in 3a)
a. Unpack the tar file into the directory of your choice (i.e. /usr/local).
b. Add the location where you installed NCBI-BLAST to your PATH variable in
.bash_profile (i.e. PATH="/usr/local/ncbi-blast:$PATH").
4. Install SNAP. Download from http://homepage.mac.com/iankorf/
a. Unpack the SNAP tar file into the directory of your choice (ie /usr/local)
b. Add the following to your .bash_profile file (value depends on where you
choose to install snap): export ZOE="/usr/local/snap/Zoe"
c. Navigate to the directory where snap was unpacked (i.e. /usr/local/snap)
and type make
d. Add the location where you installed SNAP to your PATH variable in
.bash_profile (i.e. export PATH="/usr/local/snap:$PATH").
5. Install RepeatMasker. Download from http://www.repeatmasker.org
a. The most current version of RepeatMasker requires a program called TRF.
This can be downloaded from http://tandem.bu.edu/trf/trf.html
b. The TRF download will contain a single executable file. You will need to
rename the file from whatever it is to just 'trf' (all lower case).
c. Make TRF executable by typing chmod +x+u trf. You can then move this file
wherever you want. I just put it in the /RepeatMasker directory.
d. Unpack RepeatMasker to the directory of your choice (i.e. /usr/local).
e. If you do not have WuBlast installed, you will need to install RMBlast.
We do not recomend using cross_match, as RepeatMasker performance will suffer.
f. Now in the RepeatMasker directory type perl ./configure in the command
line. You will be asked to identify the location of perl, wublast, and
trf. The script expects the paths to the folders containing the
executables (because you are pointing to a folder the path must end in a
'/' character or the configuration script throws a fit).
g. Add the location where you installed RepeatMasker to your PATH variable in
.bash_profile (i.e. export PATH="/usr/local/RepeatMasker:$PATH").
h. You must register at http://www.girinst.org and download the Repbase
repeat database, Repeat Masker edition, for RepeatMasker to work.
i. Unpack the contents of the RepBase tarball into the RepeatMasker/Libraries
directory.
6. Install Exonerate 1.4 or higher. Download from
http://www.ebi.ac.uk/~guy/exonerate
a. Exonerate has pre-comiled binaries for many systems; however, for Mac OS-X
you will have to download the source code and complile it yourself by
following steps b though d.
b. You need to have Glib 2.0 installed. The easiest way to do this on a Mac
is to install fink and then type fink install glib2-dev. For 64-bit systems
make sure fink is in 64-bit mode (which is not the default).
c. Change to the directory containing the Exonerate package to be compiled.
d. To install exonerate in the directory /usr/local/exonerate, type:
./configure -prefix=/usr/local/exonerate -> then type make -> then type
make install
e. Add the location where you installed exonerate to your PATH variable in
.bash_profile (i.e. export PATH="/usr/local/exonerate/bin:$PATH").
7. Install MAKER. Download from http://www.yandell-lab.org
a. Unpack the MAKER tar file into the directory of your choice (i.e.
/usr/local).
b. Go to the MAKER src/ directory.
c. Configure using --> perl Build.PL
D. Install using --> ./Build install
b. Remember to add the following to your .bash_profile if you haven't already:
export ZOE="where_snap_is/Zoe"
export AUGUSTUS_CONFIG_PATH="where_augustus_is/config
c. Add the location where you installed MAKER to your PATH variable in
.bash_profile (i.e. export PATH=/usr/local/maker/bin:$PATH).
d. You can now run a test of MAKER by following the instructions in the MAKER
README file.
(OPTIONAL COMPONENTS)
1. If you want to install the MPI version of MAKER, you need to have Perl
compiled for threads and install threads 1.67 or higher.
a. Install standard MAKER and verify that it runs.
b. Install MPICH2 with the --enable-sharedlibs flag set to the
appropriate value for your OS (See MPICH2 documentation)
c. Use cd to change to the maker/src subdirectory in the MAKER
instalation folder.
d. Run Build.PL by typing: perl Build.PL
e. If MPICH2 is installed and configured correctly, it will be
detected by MAKER and Build.PL will ask if you want to intall
MPI MAKER. Select 'yes'.
f. run ./Build install
2. Augustus 2.0 or higher. Download from http://augustus.gobics.de
a. Change to the directory containing the Augustus package to be compiled.
b. Unpack Augustus to the directory of your choice (i.e. /usr/local).
c. Change to the src/ directory inside the extracted augustus folder.
d. Install and compile Augustus by typing: make
e. Add the following to your .bash_profile file (value depends on where you
install augutus): export AUGUSTUS_CONFIG_PATH=/usr/local/augustus/config
f. Add the location where you installed augustus to your PATH variable in
.bash_profile (i.e. export PATH=/usr/local/augustus/bin:$PATH).
3. GeneMark-ES. Download from http://exon.biology.gatech.edu
a. See GeneMark-ES installation documentation.
4. FGENESH 2.4 or higher. Purchase from http://www.softberry.com
a. See FGENESH installation documentation.
5. GeneMarkS. Used for prokaryotic genomes only.
Download from http://exon.biology.gatech.edu
a. See GeneMarkS installation documentation.
Getting Started with MAKER
Note
Before we begin with example data I want everyone to note that there are finished examples are located in example data folder, so if you fall behind you can always find MAKER control files datasets and final results in there.
First let's test our MAKER executable and look at the usage statement:
maker -h
MAKER version 2.26
Usage:
maker [options] <maker_opts> <maker_bopts> <maker_exe>
Description:
MAKER is a program that produces gene annotations in GFF3 file format using
evidence such as EST alignments and protein homology. MAKER can be used to
produce gene annotations for new genomes as well as update annotations from
existing genome databases.
The three input arguments are user control files that specify how MAKER
should behave. All options for MAKER should be set in the control files,
but a few can also be set on the command line. Command line options provide
a convenient machanism to override commonly altered control file values.
MAKER will automatically search for the control files in the current working
directory if they are not specified on the command line.
Input files listed in the control options files must be in fasta format
unless otherwise specified. Please see MAKER documentation to learn more
about control file configuration. MAKER will automatically try and locate
the user control files in the current working directory if these arguments
are not supplied when initializing MAKER.
It is important to note that MAKER does not try and recalculated data that
it has already calculated. For example, if you run an analysis twice on
the same dataset you will notice that MAKER does not rerun any of the BLAST
analyses, but instead uses the blast analyses stored from the previous run.
To force MAKER to rerun all analyses, use the -f flag.
MAKER also supports parallelization via MPI on computer clusters. Just
launch MAKER via mpiexec (Example: mpiexec -n 40 maker). MPI support must be
configured during the MAKER installation process for this to work though.
Options:
-genome|g <file> Overrides the genome file location in the control files.
-RM_off|R Turns all repeat masking options off.
-datastore/ Forcably turn on/off MAKER's use of a two deep datastore
nodatastore directory structure for output. Always on by default.
-base <string> Set the base name MAKER uses to save output files.
MAKER uses the input genome file name by default.
-tries|t <integer> Run contigs up to the specified number of tries.
-cpus|c <integer> Tells how many cpus to use for BLAST analysis.
Note: this is for BLAST and not for MPI!
-force|f Forces MAKER to delete old files before running again.
This will require all blast analyses to be rerun.
-again|a Caculate all annotations and output files again even if
no settings have changed. Does not delete old analyses.
-quiet|q Regular quiet. Only a handlful of status messages.
-qq Even more quit. There are no status messages.
-dsindex Quickly generate datastore index file. Note that this
will not check if run settings have changed on contigs.
-CTL Generate empty control files in the current directory.
-OPTS Generates just the maker_opts.ctl file.
-BOPTS Generates just the maker_bopts.ctl file.
-EXE Generates just the maker_exe.ctl file.
-MWAS <option> Easy way to control mwas_server daemon for web-based GUI
options: STOP
START
RESTART
-version Prints the MAKER version.
-help|? Prints this usage statement.
Next let's quickly take a look at our data folders
cd ~/maker_course ls -1
You should see five example folders
example1_dmel example2_pyu example3_mRNAseq example4_legacy example5_ecoli
Let's look inside example1
ls -1 example1_dmel
You will see a directory called finished.maker.output which contains all the final results for the example. Each of the other examples will contain a similar directory.
finished.maker.output
Now let's get started!
Running MAKER with example data
MAKER comes with some example input files to test the installation and to familiarize the user with how to run MAKER. The example files are found in the maker/data directory.
ls -1 /usr/local/maker/data
dpp_contig.fasta
dpp_proteins.fasta
dpp_est.fasta
te_protein.fasta
The example files are in FASTA format. MAKER requires FASTA format for its input files. Let's take a look at one of theses files to see what the format looks like.
less /usr/local/maker/data/dpp_proteins.fasta
>dpp-CDS-5 MRAWLLLLAVLATFQTIVRVASTEDISQRFIAAIAPVAAHIPLASASGSGSGRSGSRSVG ASTSTALAKAFNPFSEPASFSDSDKSHRSKTNKKPSKSDANRQFNEVHKPRTDQLENSKN KSKQLVNKPNHNKMAVKEQRSHHKKSHHHRSHQPKQASASTESHQSSSIESIFVEEPTLV LDREVASINVPANAKAIIAEQGPSTYSKEALIKDKLKPDPSTLVEIEKSLLSLFNMKRPP KIDRSKIIIPEPMKKLYAEIMGHELDSVNIPKPGLLTKSANTVRSFTHKDSKIDDRFPHH HRFRLHFDVKSIPADEKLKAAELQLTRDALSQQVVASRSSANRTRYQVLVYDITRVGVRG QREPSYLLLDTKTVRLNSTDTVSLDVQPAVDRWLASPQRNYGLLVEVRTVRSLKPAPHHH VRLRRSADEAHERWQHKQPLLFTYTDDGRHKARSIRDVSGGEGGGKGGRNKRQPRRPTRR KNHDDTCRRHSLYVDFSDVGWDDWIVAPLGYDAYYCHGKCPFPLADHFNSTNHAVVQTLV NNMNPGKVPKACCVPTQLDSVAMLYLNDQSTVVLKNYQEMTVVGCGCR
FASTA format is fairly simple. It contains a definition line starting with '>' that contains a name for a sequence followed by the actual nucleotide or amino acid sequence on subsequent lines. The file we are looking at contains protein sequences, so the sequence uses the single letter code for amino acids.
A minimal input file set for MAKER would generally consist of a FASTA file for the genomic sequence, a FASTA file of RNA (ESTs/cDNA/mRNA transcripts) from the organism, and a FASTA file of protein sequences from the same or related organisms (or a general protein database).
If you're following this tutorial outside of the course you can copy the example files to the example1_dmel directory that is included with the MAKER distribution in the following location:
cp /path/to/maker/data/dpp* .
Next we need to tell MAKER all the details about how we want the annotation process to proceed. Because there can be many variables and options involved in annotation, command line options would be too numerous and cumbersome. Instead MAKER uses a set of configuration files which guide each run. You can create a set of generic configuration files in the current working directory by typing the following.
maker -CTL
This creates three files (type ls -1 to see).
- maker_exe.ctl - contains the path information for the underlying executables.
- maker_bopt.ctl - contains filtering statistics for BLAST and Exonerate
- maker_opt.ctl - contains all other information for MAKER, including the location of the input genome file.
Control files are run-specific and a separate set of control files will need to be generated for each genome annotated with MAKER. MAKER will look for control files in the current working directory, so it is recommended that MAKER be run in a separate directory containing unique control files for each genome.
Let's take a look at the maker_exe.ctl file.
nano maker_exe.ctl
You will see the names of a number of MAKER supported executables as well as the path to their location. If you followed the installation instructions correctly, including the instructions for installing prerequisite programs, all executable paths should show up automatically for you. However if the location to any of the executables is not set in your PATH environment variable, as per installation instructions, you will have to add these manually to the maker_exe.ctl file every time you run MAKER.
Lines in the MAKER control files have the format key=value with no spaces before or after the equals sign(=). If the value is a file name, you can use relative paths and environment variables, i.e. snap=$HOME/snap. Note that for all control files the comments written to help users begin with a pound sign(#). In addition, options before the equals sign(=) can not be changed, nor should there be a space before or after the equals sign.
Now let's take a look at the maker_bopts.ctl file.
nano maker_bopts.ctl
In this file you will find values you can edit for downstream filtering of BLAST and Exonerate alignments. At the very top of the file you will see that I have the option to tell MAKER whether I prefer to use WU-BLAST or NCBI-BLAST. We want to set this to NCBI-BLAST, since that is what is installed. We can just leave the remaining values as the default.
blast_type=ncbi+
Now let's take a look at the maker_opts.ctl file.
nano maker_opts.ctl
This is the primary configuration file for MAKER specific options. Here we need to set the location of the genome, EST, and protein input files we will be using. These come from the supplied example files. We also need to set repeat masking options, as well as a number of other configurations. We'll discuss these options in more detail later on, but for now just adjust the following values.
genome=dpp_contig.fasta est=dpp_transcripts.fasta protein=dpp_proteins.fasta est2genome=1
Note: Do not put spaces on either side of the = on the above control file lines.
Now let's run MAKER.
maker
You should now see a large amount of status information flowing past your screen. If you don't want to see this you can run MAKER with the -q option for "quiet" on future runs.
Details of What is Going on Inside of MAKER
Repeat Masking
The first step in the MAKER pipeline is repeat masking. Why do we need to do this? Repetitive elements can make up a significant portion of the genome. These repeats fall into to basic classes:
- Low-complexity (simple) repeats: These consist of stretches (sometimes very long) of tandemly repeated sequences with little information content. Examples of low-complexity sequence are mononucleotide runs (AAAAAAA, GGGGGG) and the various types of satellite DNA.
- Interspersed (complex) repeats - Sections of sequence that have the ability to change thier location within the genome. These transposons and retrotransposons contain real coding genes (reverse transcriptase, Gag, Pol) and have the ability to transpose (and often duplicate) surrounding sequence with them.
The low information content of the low complexity repeats sequence can produce sequence alignments with high statistical significance to low-complexity protein regions creating a false homology (think evidence for genes) throughout the genome.
Because these complex repeats contain real protein coding genes they play havoc with ab initio gene predictors. For example, a transposable element that occurs within the intron of one of the organism's own protein encoding genes might cause a gene predictor to include extra exons as part of this gene. Thus, sequence which really only belongs to a transposable element is included in your final gene annotation set.
Analysis of the repeat structure of a new genome is an important goal, but the presence of those repeats both simple and complex makes it nearly impossible to generate a useful annotation set of the organisms own genes. For this reason it is critical to identify and mask these repetitive regions of the genome.
MAKER identifies repeats in two steps.
- First MAKER runs a program called RepeatMasker is used to identify both all classes of repeats that match entries in the RepBase repeat library. You can even create your own species specific repeat library and RepeatMasker use it in addition to its own libraries to mask repeats based on nucleotide repeats libraries.
- Next MAKER uses RepeatRunner to identify transposable elements and viral proteins using the RepeatRunner protein database. Because RepeatRunner uses protein sequence libraries and protein sequence diverges at a slower rate than nucleotide sequence, this step picks up many problematic regions of divergent repeats that are missed by RepeatMasker (which searches in nucleotide space).
Regions identified during repeat analysis are masked out in two different ways:
- Complex repeats are hard-masked - the repeat sequence is replaced with the letter N. This essentially removes this sequence from any further consideration at any later point of the annotation process.
- Simple repeats are soft-masked - sequences are transformed to lower case. This prevents alignment programs such as Blast from seeding any new alignments in the soft-masked region, however alignments that begin in a nearby (non-masked) region of the genome can extend into the soft-masked region. This is important because low-complexity regions are found within many real genes, they just don't make up the majority of the gene.
Masking sequence from the annotation pipeline (especially hard masking) may seem like it might cause us to lose real protein coding genes that are important for the organism's biology. It is true that repeat derived genes can be co-opted and expressed by the organism and repeat masking will affect our ability to annotate these genes. However, these genes are rare and the number of gene models and sequence alignments improved by the repeat masking step far outweighs the few gene models that may be negatively affected. You do have the option to run ab initio gene predictors on both the masked and unmasked sequence if repeat masking worries you though. You do this by setting unmask:1 in the maker_opt.ctl configuration file.
Ab Initio Gene Prediction
Following repeat masking, MAKER runs ab initio gene predictors specified by the user to produce preliminary gene models. Ab initio gene predictors produce gene predictions based on underlying mathematical models describing patterns of intron/exon structure and consensus start signals. Because the patterns of gene structure are going to differ from organism to organism, you must train gene predictors before you can use them. I will discuss how to do this later on.
MAKER currently supports:
- SNAP (Works good, easy to train, not as good as others especially on longer intron genomes).
- Augustus (Works great, hard to train, but getting better)
- GeneMark (Self training, no hints, buggy, not good for fragmented genomes or long introns).
- FGENESH (Works great, costs money even for training)
You must specify in the maker_opts.ctl file the training parameters file you want to use use when running each of these algorithms.
RNA and Protein Evidence Alignment
A simple way to indicate if a sequence region is likely associated with a gene is to identify (A) if the region is actively being transcribed or (B) if the region has homology to a known protein. This can be done by aligning Expressed Sequence Tags (ESTs) and proteins to the genome using alignment algorithms.
- ESTs are sequences derived from a cDNA library. Because of the difficulties associated with working with mRNA and depending on how the cDNA library was prepared, EST databases usually represent bits and pieces of transcribed RNA with only a few full length transcripts. MAKER aligns these sequences to the genome using BLASTN. If ESTs from the organism being annotated are unavailable or sparse, you can use ESTs from a closely related organism. However, RNA from closely related organisms are unlikely to align using BLASTN since nucleotide sequences can diverge quite rapidly. For these RNAs, MAKER uses TBLASTX to align them in protein space.
- Protein sequence generally diverges quite slowly over large evolutionary distances, as a result proteins from even evolutionarily distant organisms can be aligned against raw genomic sequence to try and identify regions of homology. MAKER does this using BLASTX.
Remember now that we are aligning against the repeat-masked genomic sequence. How is this going to affect our alignments? For one thing we won't be able to align against low-complexity regions. Some real proteins contain low-complexity regions and it would be nice to identify those, but if I let anything align to a low-complexity region, then I will get spurious alignments all over the genome. Wouldn't it be nice if there was a way to allow BLAST to extend alignments through low-complexity regions, but only if there is is already alignment somewhere else? You can do this with soft-masking. If you remember soft-masking is using lower case letters to mask sequence without losing the sequence information. BLAST allows you to use soft-masking to keep alignments from seeding in low-complexity regions, but allows you to extend through them. This of course will allow some of the spurious alignments you were trying to avoid, but overall you still end up suppressing the majority of poor alignments while letting through enough real alignments to justify the cost. You can turn this behavior off though if it bothers you by setting softmask=0 in the maker_bopt.ctl file.
Polishing Evidence Alignments
Because of oddities associated with how BLAST statistics work, BLAST alignments are not as informative as they could be. BLAST will align regions any where it can, even if the algorithm aligns regions out of order, with multiple overlapping alignments in the exact same region, or with slight overhangs around splice sites.
To get more informative alignments MAKER uses the program Exonerate to polish BLAST hits. Exonerate realigns each sequences identified by BLAST around splice sites and forces the alignments to occur in order. The result is a high quality alignment that can be used to suggest near exact intron/exon positions. Polished alignments are produced using the est2genome and protein2genome options for Exonerate.
One of the benefits of polishing EST alignments is the ability to identify the strand an EST derives from. Because of amplification steps involved in building an EST library and limitations involved in some high throughput sequencing technologies, you don't necessarily know whether you're really aligning the forward or reverse transcript of an mRNA. However, if you take splice sites into account, you can only align to one strand correctly.
Integrating Evidence to Synthesize Annotations
Once you have ab initio predictions, EST alignments, and protein alignments you can integrate this evidence to produce even better gene predictions. MAKER does this by communicating with the gene prediction programs. MAKER takes all the evidence, generates "hints" to where splice sites and protein coding regions are located, and then passes these "hints" to programs that will accept them.
MAKER produces hint based predictors for:
- SNAP
- Augustus
- FGENESH
- GeneMark (under development)
Selecting and Revising the Final Gene Model
MAKER then takes the entire pool of ab initio and evidence informed gene predictions, updates features such as 5' and 3' UTRs based on EST evidence, tries to determine alternative splice forms where EST data permits, produces quality control metrics for each gene model (this is included in the output), and then MAKER chooses from among all the gene model possibilities the one that best matches the evidence. This is done using a modified sensitivity/specificity distance metric.
MAKER can use evidence from EST alignments to revise gene models to include features such as 5' and 3' UTRs.
Quality Control
Finally MAKER calculates quality control statistics to assist in downstream management and curation of gene models outside of MAKER.
MAKER's Output
If you look in the current working directory, you will see that MAKER has created an output directory called dpp_contig.maker.output. The name of the output directory is based on the input genomic sequence file, which in this case was dpp_contig.fasta.
Now let's see what's inside the output directory.
cd dpp_contig.maker.output ls -1
You should now see a list of directories and files created by MAKER.
dpp_contig_datastore dpp_contig_master_datastore_index.log maker_bopts.log maker_exe.log maker_opts.log mpi_blastdb
- The maker_opts.log, maker_exe.log, and maker_bopts.log files are logs of the control files used for this run of MAKER.
- The mpi_blastdb directory contains FASTA indexes and BLAST database files created from the input EST, protein, and repeat databases.
- The dpp_contig_master_datastore_index.log contains information on both the run status of individual contigs and information on where individual contig data is stored.
- The dpp_contig_datastore directory contains a set of subfolders, each containing the final MAKER output for individual contigs from the genomic fasta file.
Once a MAKER run is finished the most important file to look at is the dpp_contig_master_datastore_index.log to see if there were any failures.
less -S dpp_contig_master_datastore_index.log
If everything proceeded correctly you should see the following:
contig-dpp-500-500 dpp_contig_datastore/contig-dpp-500-500 STARTED contig-dpp-500-500 dpp_contig_datastore/contig-dpp-500-500 FINISHED
There are only entries describing a single contig because there was only one contig in the example file. These lines indicate that the contig contig-dpp-500-500 STARTED and then FINISHED without incident. Other possible entries include:
- FAILED - indicates a failed run on this contig, MAKER will retry these
- RETRY - indicates that MAKER is retrying a contig that failed
- SKIPPED_SMALL - indicates the contig was too short to annotate (minimum contig length is specified in maker_opt.ctl)
- DIED_SKIPPED_PERMANENT - indicates a failed contig that MAKER will not attempt to retry (number of times to retry a contig is specified in maker_opt.ctl)
The entries in the dpp_contig_master_datastore_index.log file also indicate that the output files for this contig are stored in the directory dpp_contig_datastore/contig-dpp-500-500/. Knowing where the output is stored may seem trivial, however, input genome fasta files can contain thousands even hundreds-of-thousands of contigs, and many file-systems have performance problems with large numbers of sub-directories and files within a single directory. Even when the underlying file-system handles things gracefully, access via network file-systems can still be an issue. To deal with this problem, MAKER creates a hierarchy of nested sub-directory layers, starting from a 'base', and places the results for a given contig within these datastore of possibly thousands of nested directories. The master_datastore_index.log file this is essential for identifying where the output for a given contig is stored.
Now let's take a look at what MAKER produced for the contig 'contig-dpp-500-500'.
cd dpp_contig_datastore/05/1F/contig-dpp-500-500/ ls -1
The directory should contain a number of files and a directory.
contig-dpp-500-500.gff contig-dpp-500-500.maker.proteins.fasta contig-dpp-500-500.maker.transcripts.fasta run.log theVoid.contig-dpp-500-500
- The contig-dpp-500-500.gff contains all annotations and evidence alignments in GFF3 format. This is the important file for use with Apollo or GBrowse.
- The contig-dpp-500-500.maker.transcripts.fasta and contig-dpp-500-500.maker.proteins.fasta files contain the transcript and protein sequences for MAKER produced gene annotations.
- The run.log file is a log file. If you change settings and rerun MAKER on the same dataset, or if you are running a job on an entire genome and the system fails, this file lets MAKER know what analyses need to be deleted, rerun, or can be carried over from a previous run. One advantage of this is that rerunning MAKER is extremely fast, and your runs are virtually immune to all system failures.
- The directory theVoid.contig-dpp-500-500 contains raw output files from all the programs MAKER runs (Blast, SNAP, RepeatMasker, etc.). You can usually ignore this directory and its contents.
Viewing MAKER Annotations
Let's take a look at the GFF3 file produced by MAKER.
less contig-dpp-500-500.gff
As you can see, manually viewing the raw GFF3 file produced by MAKER really isn't that meaningful. While you can identify individual features such as genes, mRNAs, and exons, trying to interpret those features in the context of thousands of other genes and thousands of bases of sequence really can't be done by directly looking at the GFF3 file.
For sanity check purposes it would be nice to have a graphical view of what's in the GFF3 file. To do that, GFF3 files can be loaded into programs like Apollo and GBrowse.
Gbrowse
Now let's look at our MAKER output in a couple ways. The AWS AMI that we are using has GBrowse set up and running, so let's have a quick look at our ourput in GBrowse. First we need to copy our 'genome database' - the GFF3 and Fasta files for this contig - to a location where GBrowser can find them. I've set up GBrowse to look in a folder in your example1_dmel folder so let's create that folder and copy our genome files there.
cd ~/maker_course/example1_dmel/ mkdir gbrowse cd ~/maker_course/example1_dmel/ cp dpp_contig.maker.output/dpp_contig_datastore/05/1F/contig-dpp-500-500/contig-dpp-500-500.gff \ ../gbrowse cp dpp_contig.fasta ./gbrowse
Now let's point our browser to our very own mini-FlyBase (fill in the appropriate AWS URL for your instance):
http://Template:AWSurl/cgi-bin/gbrowse/gbrowse/maker_dmel
Apollo
While genome browsers like Gbrowse and JBrowse are very useful for displaying and distributing our annotations to the broader scientific community, since we've created and maintain these annotations we'll want to be able to manually curate them. [Apollo] is the tool for this job!
Apollo is a desktop application not a web-application (as least as of August 2012). We could run Apollo on our AWS server to view our annotations on our laptop by setting up X-11 forwarding, but with a roomful of us running GUIS on a remote server over a shared wireless connection is asking for trouble. So let's copy our genome files to our laptop and view them there. If you don't have Apollo installed on your local machine just follow along for a while on the main screen or a neighbor's computer.
cp dpp_contig.maker.output/dpp_contig_datastore/05/1F/contig-dpp-500-500/contig-dpp-500-500.gff \ ~/maker_course/example1__dmel/gbrowse/
Your AWS connection string will be different.
Before starting Apollo let's configure it for MAKER output. MAKER comes with a configuration file that will allow Apollo to display MAKER annotations and evidence in nice color (otherwise everything will be the same color of white). Put a copy of this configuration file ~/.apollo directory.
Now start Apollo and load the contig-dpp-500-500.gff into Apollo and take a look at what MAKER produced. Copy the contig-dpp-500-500.gff file to your home directory to make it easy to locate.
cp contig-dpp-500-500.gff ~
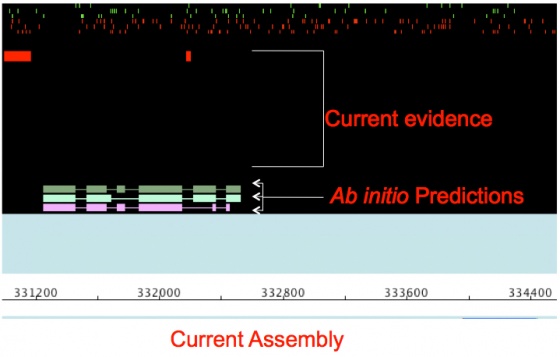
You will notice that there are a number of bars representing the gene annotations and the evidence alignments supporting those annotations. Annotations are in the middle light colored panel, and evidence alignments are in the dark panels at the top and bottom. As you have probably realized, this view is much easier to interpret than looking directly at the GFF3 file.
Now click on each piece of evidence and you will see its source in the table at the bottom of the Apollo screen.
Possible Sources Include:
- BLASTN - BLASTN alignment of EST evidence
- BLASTX - BLASTX alignment of protein evidence
- TBLASTX - TBLASTX alignment of EST evidence from closely related organisms
- EST2Genome - Polished EST alignment from Exonerate
- Protein2Genome - Polished protein alignment from Exonerate
- SNAP - SNAP ab inito gene prediction
- GENEMARK - GeneMarkab inito gene prediction
- Augustus - Augustus ab inito gene prediction
- FgenesH - FGENESH ab inito gene prediction
- Repeatmasker - RepeatMasker identified repeat
- RepeatRunner - RepeatRunner identified repeat from the repeat protein database
Advanced MAKER Configuration, Re-annotation Options, and Improving Annotation Quality
The remainder of this page addresses issues that can be encountered during the annotation process. I then describe how MAKER can be used to resolve each issue.
Configuration Files in Detail
Let's take a closer look at the configuration options in the maker_opt.ctl file.
cd /home/ubuntu/maker_course/example1_dmel nano maker_opts.ctl
Genome Options (Required)
genome=dpp_contig.fasta #genome sequence (fasta file or fasta embeded in GFF3 file) organism_type=eukaryotic #eukaryotic or prokaryotic. Default is eukaryotic
Re-annotation Using MAKER Derived GFF3
maker_gff= #MAKER derived GFF3 file est_pass=0 #use ESTs in maker_gff: 1 = yes, 0 = no altest_pass=0 #use alternate organism ESTs in maker_gff: 1 = yes, 0 = no protein_pass=0 #use protein alignments in maker_gff: 1 = yes, 0 = no rm_pass=0 #use repeats in maker_gff: 1 = yes, 0 = no model_pass=0 #use gene models in maker_gff: 1 = yes, 0 = no pred_pass=0 #use ab-initio predictions in maker_gff: 1 = yes, 0 = no other_pass=0 #passthrough anyything else in maker_gff: 1 = yes, 0 = no
RNA (EST) Evidence
est=dpp_transcripts.fasta #set of ESTs or assembled mRNA-seq in fasta format altest= #EST/cDNA sequence file in fasta format from an alternate organism est_gff= #aligned ESTs or mRNA-seq from an external GFF3 file altest_gff= #aligned ESTs from a closly relate species in GFF3 format
Protein Homology Evidence
protein=dpp_proteins.fasta #protein sequence file in fasta format (i.e. from mutiple oransisms) protein_gff= #aligned protein homology evidence from an external GFF3 file
Repeat Masking
model_org=all #select a model organism for RepBase masking in RepeatMasker rmlib= #provide an organism specific repeat library in fasta format for RepeatMasker repeat_protein=/usr/local/maker/data/te_proteins.fasta #provide # [cont'd] a fasta file of transposable element proteins for RepeatRunner rm_gff= #pre-identified repeat elements from an external GFF3 file prok_rm=0 #forces MAKER to repeatmask prokaryotes # [cont'd] (no reason to change this), 1 = yes, 0 = no softmask=1 #use soft-masking rather than hard-masking in BLAST # [cont'd] (i.e. seg and dust filtering)
Gene Prediction
snaphmm= #SNAP HMM file gmhmm= #GeneMark HMM file augustus_species= #Augustus gene prediction species model fgenesh_par_file= #FGENESH parameter file pred_gff= #ab-initio predictions from an external GFF3 file model_gff= #annotated gene models from an external GFF3 file (annotation pass-through) est2genome=1 #infer gene predictions directly from ESTs, 1 = yes, 0 = no protein2genome=0 #infer predictions from protein homology, 1 = yes, 0 = no unmask=0 #also run ab-initio prediction programs on unmasked sequence, 1 = yes, 0 = no
Other Annotation Feature Types
other_gff= #extra features to pass-through to final MAKER generated GFF3 file
External Application Behavior Options
alt_peptide=C #amino acid used to replace non-standard amino acids in BLAST databases cpus=1 #max number of cpus to use in BLAST and RepeatMasker (not for MPI, leave 1 when using MPI)
MAKER Behavior Options
max_dna_len=100000 #length for dividing up contigs into chunks (increases/decreases memory usage) min_contig=1 #skip genome contigs below this length (under 10kb are often useless) pred_flank=200 #flank for extending evidence clusters sent to gene predictors pred_stats=0 #report AED and QI statistics for all predictions as well as models AED_threshold=1 #Maximum Annotation Edit Distance allowed (bound by 0 and 1) min_protein=0 #require at least this many amino acids in predicted proteins alt_splice=0 #Take extra steps to try and find alternative splicing, 1 = yes, 0 = no always_complete=0 #extra steps to force start and stop codons, 1 = yes, 0 = no map_forward=0 #map names and attributes forward from old GFF3 genes, 1 = yes, 0 = no keep_preds=0 #Concordance threshold to add unsupported gene prediction (bound by 0 and 1) split_hit=10000 #length for the splitting of hits (expected max intron size for evidence alignments) single_exon=0 #consider single exon EST evidence when generating annotations, 1 = yes, 0 = no single_length=250 #min length required for single exon ESTs if 'single_exon is enabled' correct_est_fusion=0 #limits use of ESTs in annotation to avoid fusion genes tries=2 #number of times to try a contig if there is a failure for some reason clean_try=0 #remove all data from previous run before retrying, 1 = yes, 0 = no clean_up=0 #removes theVoid directory with individual analysis files, 1 = yes, 0 = no TMP= #specify a directory other than the system default temporary directory for temporary files
Training ab initio Gene Predictors
If you are involved in a genome project for an emerging model organism, you should already have an EST database which would have been generated as part of the original sequencing project. A protein database can be collected from closely related organism genome databases or by using the UniProt/SwissProt protein database or the NCBI NR protein database. However a trained ab initio gene predictor is a much more difficult thing to generate. Gene predictors require existing gene models on which to base prediction parameters. However, with emerging model organisms you are not likely to have any pre-existing gene models. So how then are you supposed to train your gene prediction programs?
MAKER gives the user the option to produce gene annotations directly from the EST evidence. You can then use these imperfect gene models to train gene predictor program. Once you have re-run MAKER with the newly trained gene predictor, you can use the second set of gene annotations to train the gene predictors yet again. This boot-strap process allows you to iteratively improve the performance of ab initio gene predictors.
I've created an example file set so you can learn to train the gene predictor SNAP using this procedure.
First let's move to the example directory.
cd /home/ubuntu/maker_course/example2_pyu ls -1
You should see the following files (plus others) in the directory
pyu-contig.fasta pyu-est.fasta pyu-protein.fasta
We need to build maker configuration files and populate the appropriate values.
maker -CTL nano maker_opts.ctl
Edit the following:
genome=pyu-contig.fasta est=pyu-est.fasta protein=pyu-protein.fasta est2genome=1
MAKER is now configured to generate annotations from the EST data, so start the program (this will take a minute to run).
maker
Once finished load the file pyu-contig.maker.output/pyu-contig_datastore/scf1117875581239.gff into Apollo. You will see that there are far more regions with evidence alignments than there are gene annotations. This is because there are so few spliced ESTs that are capable of generating gene models.
Now exit Apollo. We now need to convert the GFF3 gene models to ZFF format. This is the format SNAP requires for training. To do this wee need to collect all GFF3 files into a single directory.
mkdir snap cp pyu-contig.maker.output/pyu-contig_datastore/09/14/scf1117875582023/scf1117875582023.gff snap/ cd snap maker2zff scf1117875582023.gff ls -1
There should now be two new files. The first is the ZFF format file and the second is a FASTA file the coordinates can be referenced against. These will be used to train SNAP.
genome.dna
The basic steps for training SNAP are first to filter the input gene models, then capture genomic sequence immediately surrounding each model locus, and finally uses those captured segments to produce the HMM. You can explore the internal SNAP documentation for more details if you wish.
fathom -categorize 1000 genome.ann genome.dna fathom -export 1000 -plus uni.ann uni.dna forge export.ann export.dna hmm-assembler.pl Pult . > Pult.hmm cd ..
The final training parameters file is Pult.hmm. We do not expect SNAP to perform that well with this training file because it is based on incomplete gene models; however, this file is a good starting point for further training.
We need to run MAKER again with the new HMM file we just built for SNAP.
nano maker_opts.ctl
And set:
snaphmm=snap/Pult.hmm est2genome=0
And run
maker
Now let's look at the output in Apollo. When you examine the annotations you should notice that final MAKER gene models displayed in light blue, are more abundant now and are in relatively good agreement with the evidence alignments. However the SNAP ab initio gene predictions in the evidence tier do not yet match the evidence that well. This is because SNAP predictions are based solely on the mathematic descriptions in the HMM; whereas, MAKER models also use evidence alignments to help further inform gene models. This demonstrates why you get better performance by running ab initio gene predictors like SNAP inside of MAKER rather than producing gene models by themselves for emerging model organism genomes. The fact that the MAKER models are in better agreement with the evidence than the current SNAP models also means I can use the MAKER models to retrain SNAP in a bootstrap fashion, thereby improving SNAP's performance and consequentially MAKER's performance.
Close Apollo, retrain SNAP, and run MAKER again.
mkdir snap2 cp pyu-contig.maker.output/pyu-contig_datastore/scf1117875582023/scf1117875582023.gff snap2/ cd snap2 maker2zff scf1117875582023.gff fathom -categorize 1000 genome.ann genome.dna fathom -export 1000 -plus uni.ann uni.dna forge export.ann export.dna hmm-assembler.pl Pult . > Pult2.hmm cd .. nano maker_opts.ctl
Change configuration file.
snaphmm=snap2/Pult2.hmm
Run maker.
maker
Let's examine the GFF3 file one last time in Apollo. As you can see there, there is now a marked degree of improvement in both the MAKER and SNAP gene models, and both models are in more agreement with each other.
MAKER Web Annotation Service
As you have all experienced with the previous examples, running programs on the command line can seem difficult. Many users might feel overwhelmed by trying to install and run a program like MAKER locally, especially if they are not very familiar with Linux. For those individuals, our lab has produced the MAKER Web Annotation Service (MWAS). MWAS is a website where you can run MAKER over the web without having to install any software locally, and you are provided with a much more user friendly interface for configuring MAKER and viewing results.
- Go to http://www.yandell-lab.org and select MWAS from the tabbed menu. You will see a link at the bottom of the page to access the MAKER Web Annotation Server. On the MWAS server page log in as a guest, then select 'New Job' from the top of the page.
Scrolling down the page, you should notice there are options to select the genome file, EST and protein evidence files, and choose ab initio gene predictors. At the top of the page select Example Jobs → D. melanogaster :Dpp' and click 'Load.
Now if you scroll down, you should notice that the values for your genome, EST and protein files has been filled out for you. At the bottom of the page click Add Job to Queue. You will now be sent to the job status page.
You will need to click Refresh Job Status, a couple of times until your job finishes. When your job is finished you will see an icon in the column marked Log. Click it. A window will come up displaying any errors that occurred for your job, so ideally this window will be blank. Next click on the View Results icon.
The results window will provide a brief summary of the status of each contig in your job, and will give you the opportunity to download the data, or view the results for individual contigs. Click on View in Apollo. This will open your data in Apollo (Ed Lee will describe just how launching Apollo over the web works during the Apollo section). Then close Apollo and click on SOBA statistics. This will open up a tool from the Sequence Ontology Consortium that provides simple summary statistics of features in a GFF3 file.
mRNAseq
mRNAseq is a high throughput technique for sequencing the entire transcriptome, and it holds the promise of allowing researchers to identify all exons and alternative splice forms for every gene in the genome with a single experiment. It may soon make gene predictors (mostly) a thing of the past.
- Still need to de-convolute reads & evidence (for now)
- Still need to archive, manage, and distribute annotations
By mapping mRNAseq reads using programs like TopHat and Bowtie, you can create GFF3 files of read islands and junctions. This data can then be passed in as EST evidence and will be used for generating hint based gene prediction and for choosing final annotations.
Load example on MWAS site. http://derringer.genetics.utah.edu/MWAS/
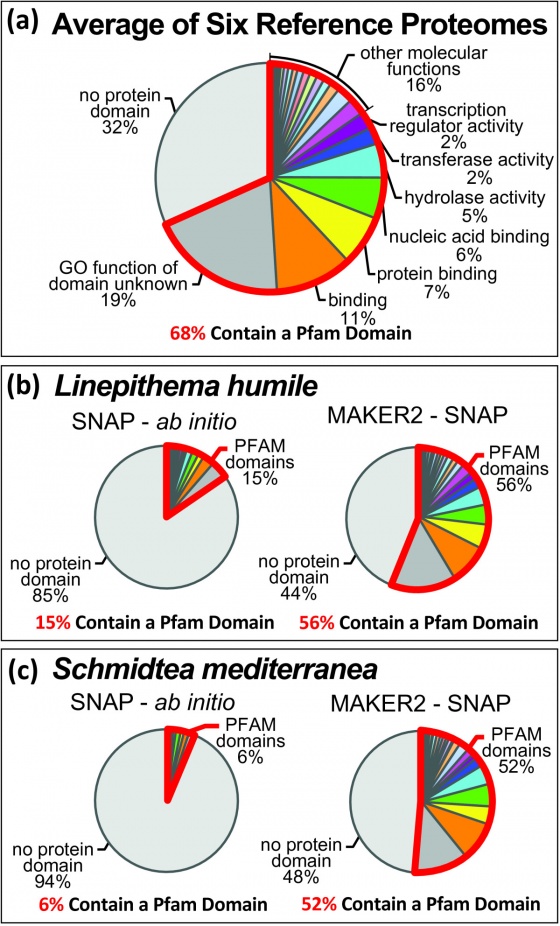
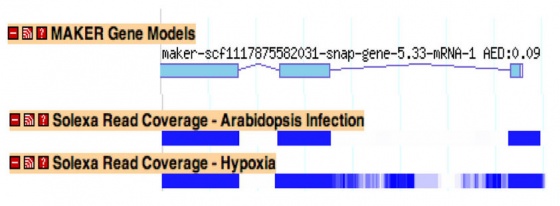
Improving Annotation Quality with MAKER's AED score
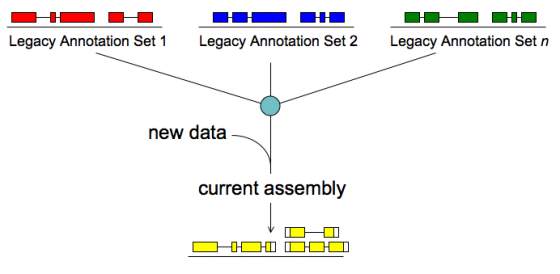
Merge/Resolve Legacy Annotations
Legacy annotations
- Many are no longer maintained by original creators
- In some cases more than one group has annotated the same genome, using very different procedures, even different assemblies
- Many investigators have their own genome-scale data and would like a private set of annotations that reflect these data
- There will be a need to revise, merge, evaluate, and verify legacy annotation sets in light of RNA-seq and other data
MAKER will:
- Identify legacy annotation most consistent with new data
- Automatically revise it in light of new data
- If no existing annotation, create new one
Load example on MWAS class site. http://derringer.genetics.utah.edu/MWAS/
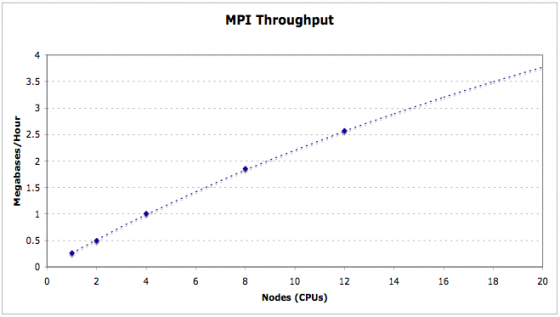
MPI Support
MAKER optionally supports Message Passing Interface (MPI), a parallel computation communication protocol primarily used on computer clusters. This allows MAKER jobs to be broken up across multiple nodes/processors for increased performance and scalability.
To use this feature, you must have MPICH2 installed with the the --enable-sharedlibs flag set during installation (See MPICH2 Installer's Guide). I have installed this for you. So let's set up MPI_MAKER and run the example file that comes with MAKER.
cd /usr/local/maker/src perl Build.PL
Accept the default that we want to build for MPI support
./Build install
You should now see the executable mpi_maker listed among the MAKER scripts (/maker/bin). Let's run some example data to see if MPI_MAKER is working properly.
cd ~ mkdir ~/maker_run2 cd maker_run2 cp /usr/local/maker/data/dpp_* ~/maker_run2 maker -CTL nano maker_opts.ctl
Set values in maker configuration files.
genome=dpp_contig.fasta est=dpp_est.fasta protein=dpp_protein.fasta snap=/usr/local/maker/exe/snap/HMM/fly
We need to set up a few more things for MPI to work. Type mpd to see a list of instructions.
mpd
You should see the following.
configuration file /home/ubuntu/.mpd.conf not found A file named .mpd.conf file must be present in the user's home directory (/etc/mpd.conf if root) with read and write access only for the user, and must contain at least a line with: MPD_SECRETWORD=<secretword> One way to safely create this file is to do the following: cd $HOME touch .mpd.conf chmod 600 .mpd.conf and then use an editor to insert a line like MPD_SECRETWORD=mr45-j9z into the file. (Of course use some other secret word than mr45-j9z.)
Follow the instructions to set this file up, and start the mpi environment with mpdboot. Then run mpi_maker through the MPI manager mpiexec.
mpdboot mpiexec -n 2 mpi_maker
mpiexec is a wrapper that handles the MPI environment. The -n 2 flag tells mpiexec to use 2 cpus/nodes when running mpi_maker. For a large cluster, this could be set to something like 100. You should now know how to start a MAKER job via MPI.
User Interface for Local MAKER Instalation
This example did not work during class because a conflict with the version of Apache that was installed. The issue has since been fixed. Before beginning the example, open a terminal and remove the following files otherwise the subversion update of maker fails.
rm ~/Documents/Software/maker/MWAS/bin/mwas_server rm ~/Documents/Software/maker/MWAS/cgi-bin/tt_templates/apollo_webstart.tt
Then update maker via subversion.
svn update ~/Documents/Software/maker/
The MWAS interface provides a very convenient method for running MAKER and viewing results; however, because compute resources are limited users are only allowed to submit a maximum of 2 megabases of sequence per job. So while MWAS might be suitable for some analyses (i.e. annotating BACs and short preliminary assemblies), if you plan on annotating an entire genome you will need to install MAKER locally. But if you like the convenience of the MWAS user interface, you can optionally install the interface on top of a locally installed version of MAKER for use in your own lab.
First under the maker directory there is a subdirectory called MWAS. MWAS contains all the needed files to build the MAKER web interface. The maker/MWAS/bin/mwas_server file is used to setup and run this web interface. Let's configure that now. There are three steps to setting up the server. First you must create and edit a server configuration file, then load all other configuration files, and then install all files to the appropriate web accessible directory.
cd /home/gmod/Documents/Software/maker/MWAS/ bin/mwas_server PREP
This will create a file in /maker/MWAS/config/ called server.ctl. We will need to edit this file before continuing.
nano config/server.ctl
Set:
apache_user:www-data web_address:http://localhost cgi_dir:/usr/lib/cgi-bin/maker cgi_web:/cgi-bin/maker html_dir:/var/www/maker html_web:/maker data_dir:/var/www/maker/data use_login:0
Now we need to generate other settings that are dependent on the values in
server_opts.ctl.
bin/mwas_server CONFIG
Several new configuration files should now be loaded in the config/ directory. These new files define default MAKER options for the server and the location of files for the server dropdown menus.
maker_bopts.ctl maker_exe.ctl maker_opts.ctl menus.ctl
We shouldn't need to edit any of these file. So let's copy files to the appropriate web accessible directories. This must be done as root or using sudo.
sudo bin/mwas_server SETUP
If you set APOLLO_ROOT in the server.ctl file, then you can now setup a special Java Web Start version of Apollo to view results directly from the web interface. Web Start will be described in more detail in the Apollo session. This must be done as root or using sudo.
sudo bin/mwas_server APOLLO
We can now run MAKER examples using this web interface, but first we need to launch a server to monitor for new job submissions.
sudo bin/mwas_server START
And then go to
MAKER Accessory Scripts
MAKER comes with a number of accessory scripts that assist in manipulations of the MAKER input and output files.
Scripts:
- add_utr_start_stop_gff - Adds explicit 5' and 3' UTR as well as start and stop codon features to the GFF3 output file
add_utr_start_stop_gff <gff3_file>
- add_utr_to_gff3.pl - Adds explicit 5' and 3' UTR features to the GFF3 output file
add_utr_gff.pl <gff3_directory>
- cegma2zff' - This script converts the output of a GFF file from CEGMA into ZFF format for use in SNAP training. Output files are always genome.ann and genome.dna
cegma2zff <cegma_gff> <genome_fasta>
- chado2gff3 - This script takes default CHADO database content and produces GFF3 files for each contig/chromosome.
chado2gff3 [OPTION] <database_name>
- compare - This script compares the contents of a GFF3 file to a CHADO database to look for merged, split and missing genes.
compare [OPTION] <database_name> <gff3_file>
- cufflinks2gff3 - This script converts the cufflinks output transcripts.gtf file into GFF3 format for use in MAKER via GFF3 passthrough. By default standless features which correspond to single exon cufflinks models will be ignored. This is because these features can correspond to repetative elements and pseudogenes. Ouput is to STDOUT so you will need to redirect to a file.
cufflinks2gff3 <transcripts1.gtf> <transcripts2.gtf> ...
- evaluator - Evaluate the the quality of an annotation set.
mpi_evaluator [options] <eval_opts> <eval_bopts> <eval_exe>
- fasta_merge - Collects all of MAKER's fasta file output for each contig and merges them to make genome level fastas
fasta_merge -d <datastore_index> -o <outfile>
- fasta_tool - The script can search, reformat, and manipulate a fasta file in a variety of ways.
- fix_fasta - Deprecated, use fasta_tool
- genemark_gtf2gff3 - This converts genemark's GTF output into GFF3 format. The script prints to STDOUT. Use the '>' character to redirect output into a file.
genemark_gtf2gff3 <filename>
- gff3_2_gtf - Converts MAKER GFF3 files to GTF format (run add_utr_start_stop_gff first to get UTR features)
gff3_2_gtf <gff3_file>
- gff3_merge - Collects all of MAKER's GFF3 file output for each contig and merges them to make a single genome level GFF3
gff3_merge -d <datastore_index> -o <outfile>
- gff3_preds2models - Converts the gene prediction match/match_part format to annotation gene/mRNA/exon/CDS format
gff3_preds2models <gff3 file> <pred list>
- gff3_to_eval_gtf - This script converts MAKER GFF3 files into GTF formated files for the program EVAL (an annotation sensitivity/specificity evaluating program). The script will only extract features explicitly declared in the GFF3 file, and will skip implicit features (i.e. UTR, start codons, and stop codons). To extract implicit features to the GTF file, you will first need to expicitly declare them in the GFF3 file. This can be done by calling the script add_utr_to_gff3 to add formal declaration lines to the GFF3 file.
gff3_to_eval_gtf <maker_gff3_file>
- iprscan2gff3 - Takes InerproScan (iprscan) output and generates GFF3 features representing domains. Interesting tier for GBrowse.
iprscan2gff3 <iprscan_file> <gff3_fasta>
- iprscan_batch - Wrapper for iprscan to take advantage of multiprocessor systems.
iprscan_batch <file_name> <cpus> <log_file>
- iprscan_wrap - A wrapper that will run iprscan
- ipr_update_gff - Takes InterproScan (iptrscan) output and maps domain IDs and GO terms to the Dbxref and Ontology_term attributes in the GFF3 file.
ipr_update_gff <gff3_file> <iprscan_file>
- maker2chado - This script takes MAKER produced GFF3 files and dumps them into a Chado database. You must set the database up first according to CHADO installation instructions. CHADO provides its own methods for loading GFF3, but this script makes it easier for MAKER specific data. You can either provide the datastore index file produced by MAKER to the script or add the GFF3 files as command line arguments.
maker2chado [OPTION] <database_name> <gff3file1> <gff3file2> ...
- maker2jbrowse - This script will produce a JBrowse data set from MAKER gff3 files.
maker2chado [OPTION] <database_name> <gff3file1> <gff3file2> ...
- maker2zff.pl - Pulls out MAKER gene models from the MAKER GFF3 output and convert them into ZFF format for SNAP training.
maker2zff.pl <gff3_file>
- maker_functional
- maker_functional_fasta - Maps putative functions identified from BLASTP against UniProt/SwissProt to the MAKER produced tarnscript and protein fasta files.
maker_functional_fasta <uniprot_fasta> <blast_output> <fasta1> <fasta2> <fasta3> ...
- maker_functional_gff - Maps putative functions identified from BLASTP against UniProt/SwissProt to the MAKER produced GFF3 files in the Note attribute.
maker_functional_gff <uniprot_fasta> <blast_output> <gff3_1>
- maker_map_ids - Build shorter IDs/Names for MAKER genes and transcripts following the NCBI suggested naming format.
maker_map_ids --prefix PYU1_ --justify 6 genome.all.gff > genome.all.id.map
- map2assembly - Maps old gene models to a new assembly where possible.
map2assembly <genome.fasta> <transcripts.fasta>
- map_data_ids - This script takes a id map file and changes the name of the ID in a data file. The map file is a two column tab delimited file with two columns: old_name and new_name. The data file is assumed to be tab delimited by default, but this can be altered with the delimit option. The ID in the data file can be in any column and is specified by the col option which defaults to the first column.
map_data_ids genome.all.id.map data.txt
- map_fasta_ids - Maps short IDs/Names to MAKER fasta files.
map_fasta_ids <map_file> <fasta_file>
- map_gff_ids - Maps short IDs/Names to MAKER GFF3 files, old IDs/Names are mapped to to the Alias attribute.
map_gff_ids <map_file> <gff3_file>
- split_fasta - Splits multi-fasta files into the number of files specified by the user. Useful for breaking up MAKER jobs.
split_fasta [count] <input_fasta>
- tophat2gff3 - This script converts the juctions file producted by TopHat into GFF3 format for use with MAKER.
tophat2gff3 <junctions.bed>